Building Baseline RAG Pipelines with OpenAI and LLAMA 3 8B from Scratch
This is the first post in a series about advanced retrieval-augmented generation (RAG) applications and how you can use them to interface with your custom data.
We will briefly introduce RAG and provide resources for further learning. Then, we will build two basic RAG pipelines that will be used in later posts to illustrate failure modes, advanced techniques to overcome them, and evaluate improvements using a validation dataset.
Basics of RAG: Why it is useful
RAG, or retrieval-augmented generation, is a method that enhances the capabilities of language models by integrating a retrieval component [1]. This allows the system to fetch relevant information from a database or document collection and use it to generate more accurate and contextually relevant responses. RAG is particularly useful for applications requiring precise and up-to-date information, such as research paper summarization and topic-based quizzing.
Three Components of RAG
A basic RAG system consists of three components.
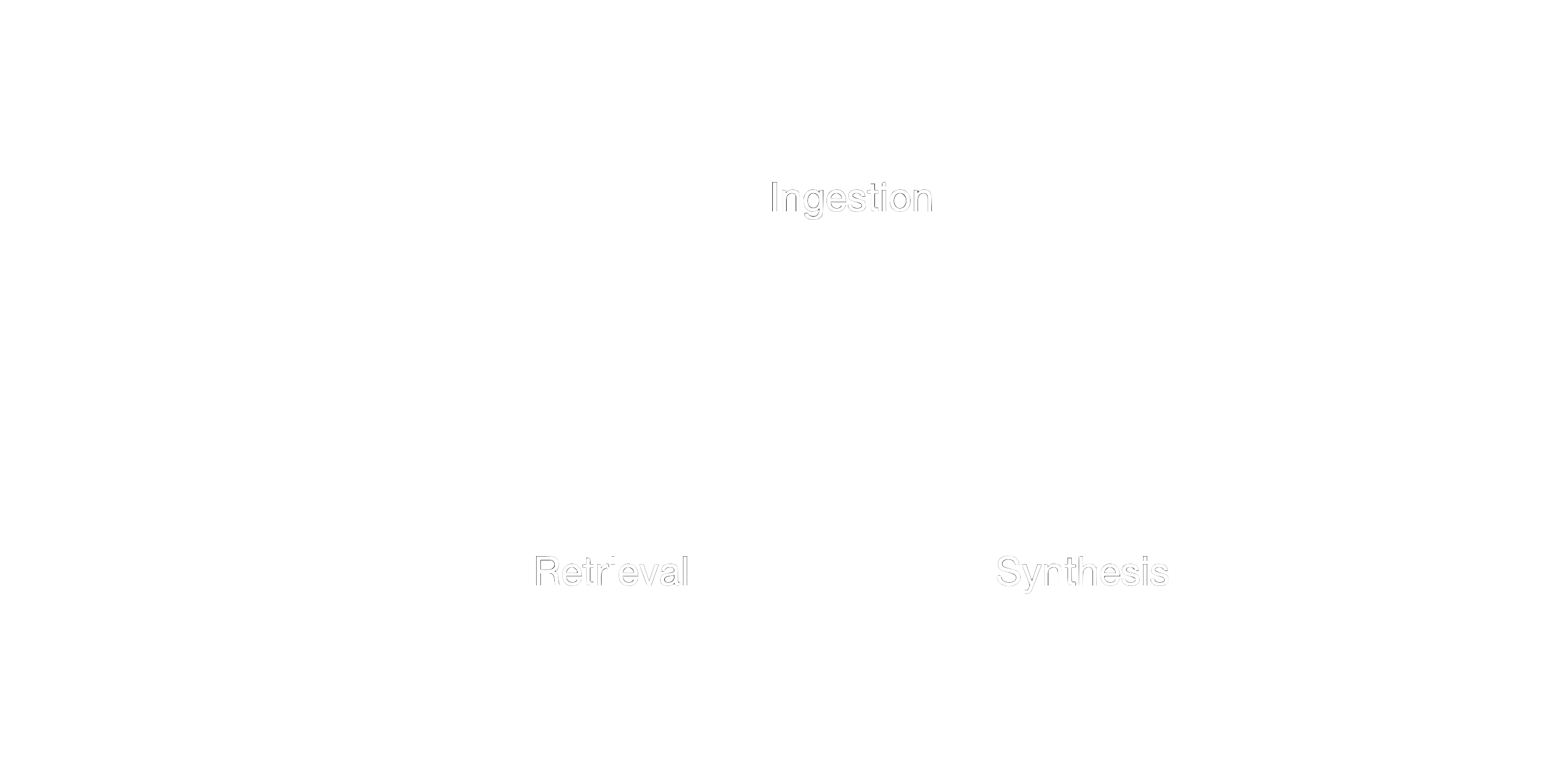
Figure 1: Three components of a RAG system.
Ingestion: The ingestion phase involves collecting and preparing data for the retrieval component. This can include a variety of data sources, such as academic papers, internal documents, or any other relevant text. The data is processed and indexed to facilitate efficient and accurate retrieval later on.
Retrieval: During the retrieval phase, the system searches through the ingested data to find the most relevant information. This is typically done using embeddings and similarity search techniques. The goal is to identify the pieces of information that are most relevant to the query or task at hand.
Synthesis/Generation: The synthesis phase involves using the retrieved information to generate a coherent and contextually appropriate response. This is where the language model comes into play, combining the retrieved data with its own knowledge to produce a final output that is both informative and relevant.
Designing Baseline RAG Pipelines
Let's develop two basic RAG pipelines: one cloud-based and one local. We will expand on these in later posts where we introduce advanced RAG techniques and use them to conduct experiments about RAG failure modes, evaluation metrics, and the impact of advanced techniques.
When making the code I decided to make it as modular as possible at the cost of having it seem more complex. That is why I will not show it here in detail, but instead will explain the architecture and reasons behind the choices. You can explore the code in the repository. Check out the tag baselines if you want to see the baseline pipelines only.
To verify if the system works we will for now simply see if the RAG paper is ingested, and if we get a useful answer to a question. We will look into proper evaluation of pipelines in future posts.
Requirements
To conduct experiments effectively, we need to ensure our code is modular and easily changeable. Additionally, it should allow us to document individual experiments to keep track of changes and their impacts on the pipelines. Here are the specific requirements around which the code is developed:
Modularity: The code should be organized into separate modules (ingestion, retrieval, synthesis) to facilitate easy adjustments and enhancements.
Configuration Management: Use a configuration file to store settings and parameters for running experiments. This makes it easy to tweak settings without changing the code.
Prompt Management: Define a file to store the prompt template and a list of queries. This ensures consistency and reproducibility across experiments.
Result Tracking: Store the results of each experiment in a JSON file, including relevant information about each run to track performance and changes over time.
Scalability: The system should be capable of handling large datasets, ensuring efficient ingestion, retrieval, and synthesis processes.
Compatibility: Ensure compatibility with different models and databases to allow comparisons between cloud-based and local pipelines.
Architecture
To meet our defined requirements and allow for efficient tracking and versioning of experiments, we structure our RAG system with modular components. This section outlines the architecture and the rationale behind our choices.
We introduce two main files to manage configuration and queries: config.yaml and prompt_queries.json. These files help us maintain consistency and reproducibility across different experiments.
Configuration Management: We use the config.yaml file to store all relevant settings and parameters for running experiments. This file allows us to easily adjust the settings without altering the core code. Key settings include paths to data sources, model parameters, and database configurations.
Prompt Management: The prompt_queries.json file contains the prompt template and a list of queries. This ensures that each experiment uses a consistent set of prompts and queries, making it easier to compare results across different runs.
Result Tracking: We store the results of each experiment in a JSON file, which includes relevant information about the specific run, such as the settings used and the outcomes. This allows us to track performance and changes over time, ensuring we can evaluate the impact of different modifications.
With these interfaces defined, let's design the three components of the RAG system.
Figure 2 shows the details of the ingestion component. Some of the steps are shared by all pipelines, while others are pipeline-specific. Overall, it has two main tasks:
Data Loading and Splitting: Regardless of the chosen pipeline (cloud-based or local), we load and split documents in a uniform manner to enable fair performance comparisons. By doing so, we must keep in mind that our maximum chunk size is limited by the smallest embedding amongst all pipelines.
Embedding Creation: We use different embedding models in each pipeline to create embeddings for the document chunks. These embeddings are then ingested into the respective databases.
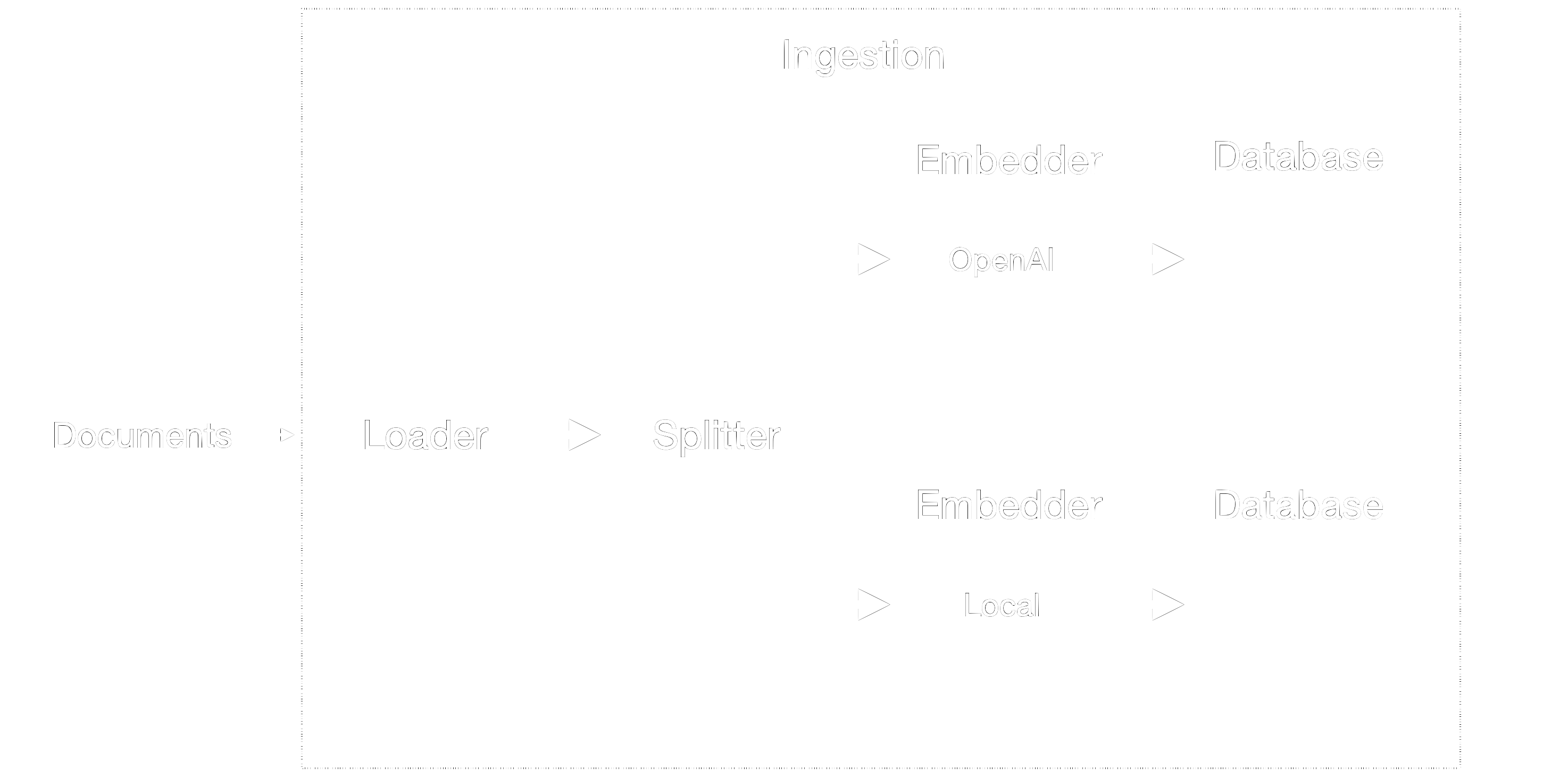
Figure 2: Ingestion component.
The retrieval and synthesis components are different for each pipeline as shown in figure 3. It has the following tasks:
- Query Embedding: Given a query, we first create an embedding using the appropriate model.
- Database Querying: The query embedding is used to search the database and retrieve the most relevant text chunks.
- Prompt Construction: The retrieved text chunks are combined with the query to create a prompt template.
- Response Generation: The prompt is sent to the language model to generate a response.
- Result Saving: The response, along with other relevant information, is saved to disk for analysis.
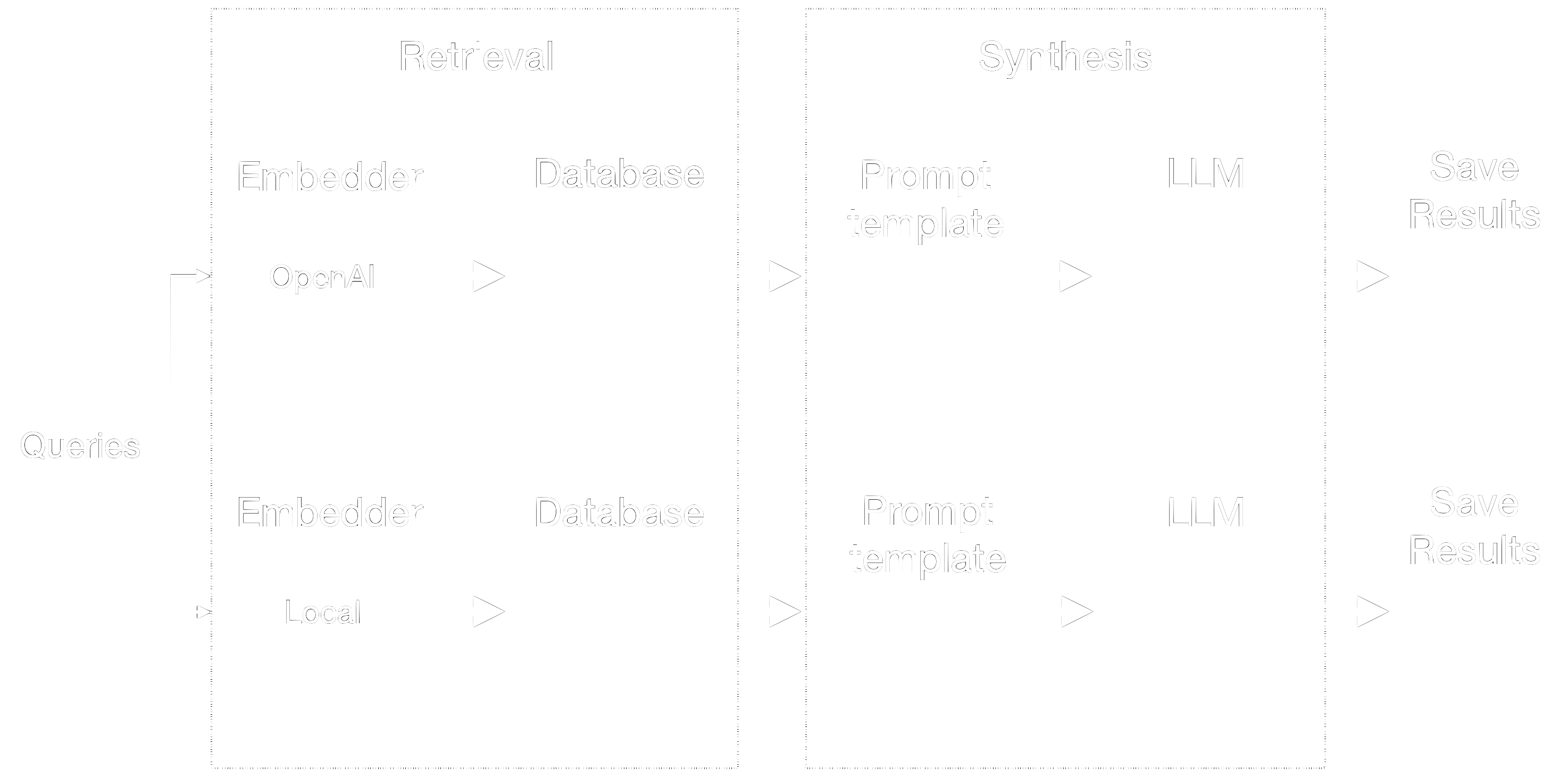
Figure 3: Retrieval and Synthesis components.
Implementing Baseline RAG Pipelines
The implementation details of our RAG system are documented in the repository, but here is a brief overview of the key libraries and decisions made during development. Keep in mind that they might change.
PDF Parsing: We use PyPDF as a parser to handle the ingestion of PDFs. This allows us to extract text documents efficiently.
Document Splitting: For splitting documents into manageable chunks, we use Langchain's Recursive splitter. Langchain's flexibility allows us to easily swap in other splitters if needed.
Database: We utilize Chroma Database due to its simplicity and effectiveness. Each pipeline has its own collection within Chroma, making it easy to manage and query the data.
OpenAIPipeline: This pipeline uses OpenAI's for both embedding and LLM tasks. The OpenAI API provides robust and reliable performance for our cloud-based solution.
LocalPipeline: For our local solution, we use LLAMA 3 8B through OLLAMA as the language model and Sentence Transformer for embedding. This setup is particularly useful for handling sensitive data and minimizing latency.
Model Objects: We use Python's
dataclassesto define model objects. These objects help us manage the result objects and other entities in a structured and type-safe manner.
By leveraging these libraries and tools, we ensure that our RAG system is modular, scalable, and easy to extend for future experiments. For detailed implementation steps and setup instructions, please refer to the repository.
Verifying the pipelines
To verify if the pipeline works correctly, we can ingest the paper. To do so, we add the path to it to the config.yaml and run python run_ingestion.py. Then, we can run python run_experiments.py with the prompt and query defined in the queries.json.
Conclusion
In this post, we introduced the basics of retrieval-augmented generation (RAG) and highlighted its importance in enhancing language models by integrating retrieval components. We outlined the three key components of a RAG system: ingestion, retrieval, and synthesis, and explained how each contributes to generating more accurate and contextually relevant responses.
We also designed and implemented two baseline RAG pipelines: one cloud-based using OpenAI's services, and one local using LLAMA 3 8B and Sentence Transformer. By making the code modular and leveraging powerful libraries like Langchain for document splitting and the Chroma Database for data management, we ensured that our system is flexible and scalable for various experiments.
Our implementation focuses on simplicity and extensibility, using dataclasses for model objects and ensuring compatibility with different models and databases. This design will allow us to explore advanced techniques and improve RAG performance in future posts. In the next posts, we will delve into the failure modes of RAG pipelines, discuss how to evaluate their performance, and explore advanced methods to address specific weaknesses. Stay tuned for detailed experiments and analyses to further enhance the capabilities of RAG systems.
References
[1] Patrick Lewis et al. (2020) Retrieval-Augmented Generation for Knowledge-Intensive NLP Tasks https://arxiv.org/abs/2005.11401
Like thinking in systems?
Subscribe here to see how I apply the same engineering mindset to building a business.
Loading comments...
Loading user status...